[% INCLUDE header.us3
title = 'UltraScan III Model Associations'
%]
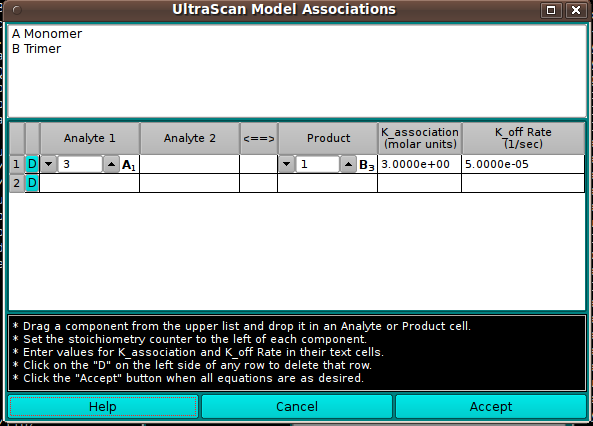
Model Associations:
In this dialog, you can define reversible associations of model
components. Associations are constructed in terms of chemical equations
with one or two analytes on one side and a product on the other.
Each association row is built by dragging a model component in the
upper list to an Analyte or Product cell. As each analyte is dropped, its
code letter (A, B, C, ...) appears in the cell with a subscript that
represents the oligomer number of the molecule. At least one Analyte and
one Product cell must be thus populated. The implied chemical equation
must be balanced by setting the stoichiometry counter in each cell such
that stoichiometry times oligomer value(s) on the left equals the product
of stoichiometry and oligomer on the right. K_association and K_off Rate
values should be entered for each row.
Once all association rows have been validly built, click the Accept
button to set the reversible association parameters for the model. If a
row is wrong and cannot be easily corrected, click the "D" button on the
left side of the row to delete that row so that it can be rebuilt.
Dialog Items:
- (analyte list) The upper list of model components is populated
from the model passed by the calling object. Members of this list can
be dragged to the table below to construct association equations.
- (chemical equation associations table) A table with one
or more rows is constructed to represent the reversible associations
present in the model.
- D These buttons in the first table column allow you to
delete the associated row.
- Analyte 1 Each row for an association must have this
column populated by dragging and dropping a list component.
- Analyte 2 Each row for an association may optionally have this
column populated by dragging and dropping a list component.
- Product Each row for an association must have this
column populated by dragging and dropping a list component. Like the
Analyte cell(s), a component will be represented by its code letter
and a subscript showing its oligomer number. The stoichiometry
counter should be set so stoichiometry-times-oligomer balances on
the sides of the equation.
- K_association Enter the K_association value in molar units
for each association row.
- K_off Rate Enter the K_off Rate value in reciprocal seconds
for each association row.
- Help Show this documentation.
- Cancel Close the dialog and do not return association
specifications to the caller.
- Accept Close the dialog and return association
specifications to the caller.
If you click on the "Accept" button and any equation does not balance,
a dialog informing you of the imbalance will pop up. You then have the
opportunity to correct the equation and "Accept" again or to click on
the "Cancel" button.
[% INCLUDE footer.us3 %]
 Manual
Manual
 Manual
Manual
